Live-dead assay on unlabeled cells using phase imaging with computational specificity
Existing approaches to evaluate cell viability involve cell staining with chemical reagents. However, the step of exogenous staining makes these methods undesirable for rapid, nondestructive, and long-term investigation. Here, we present an instantaneous viability assessment of unlabeled cells using phase imaging with computation specificity. This concept utilizes deep learning techniques to compute viability markers associated with the specimen measured by label-free quantitative phase imaging. Demonstrated on different live cell cultures, the proposed method reports approximately 95% accuracy in identifying live and dead cells. The evolution of the cell dry mass and nucleus area for the labeled and unlabeled populations reveal that the chemical reagents decrease viability. The nondestructive approach presented here may find a broad range of applications, from monitoring the production of biopharmaceuticals to assessing the effectiveness of cancer treatments.
Publications
1. Hu, C., He, S., Lee, Y. J., He, Y., Kong, E. M., Li, H., … & Popescu, G. (2022). Live-dead assay on unlabeled cells using phase imaging with computational specificity. Nature Communications, 13(1), 1-8.
3D microscopy and deep learning reveal the heterogeneity of crown-like structure microenvironments in intact adipose tissue
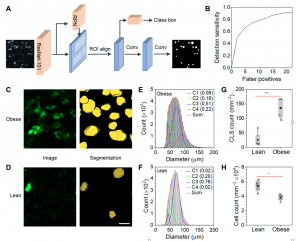
Crown-like structures (CLSs) are adipose microenvironments of macrophages engulfing adipocytes. Their histological density in visceral adipose tissue (VAT) predicts metabolic disorder progression in obesity and is believed to initiate obesity comorbidities. Here, we use three-dimensional (3D) light sheet microscopy and deep learning to quantify 3D features of VAT CLSs in lean andobese states. Obese CLS densities are significantly higher, composing 3.9% of tissue volume compared with 0.46% in lean tissue. Across the states, individual CLS structural characteristics span similar ranges; however, subpopulations are distinguishable. Obese VAT contains large CLSs absent from lean tissues, located near the tissue center, while lean CLSs have higher volumetric cell densities and prolate shapes. These features are consistent with inefficient adipocyte elimination in obesity that contributes to chronic inflammation, representing histological biomarkers to assess adipose pathogenesis. This tissue processing, imaging, and analysis pipeline can be applied to quantitatively classify 3D microenvironments across diverse tissues.
Publications
1. Geng, J., Zhang, X., Prabhu, S., Shahoei, S. H., Nelson, E. R., Swanson, K. S., … & Smith, A. M. (2021). 3D microscopy and deep learning reveal the heterogeneity of crown-like structure microenvironments in intact adipose tissue.Science Advances,7(8), eabe2480.
Image-to-image translation of label free microscopy images
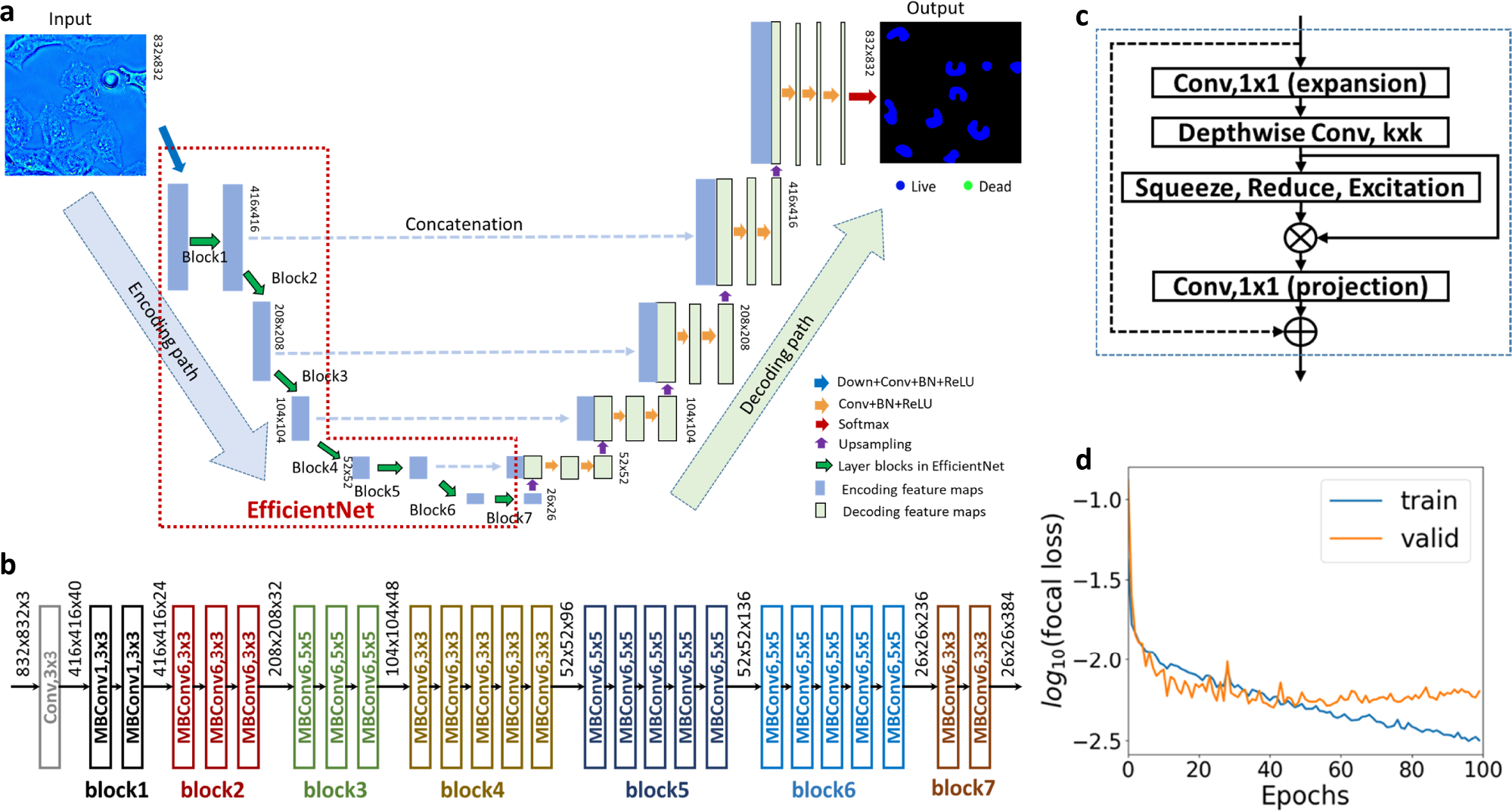
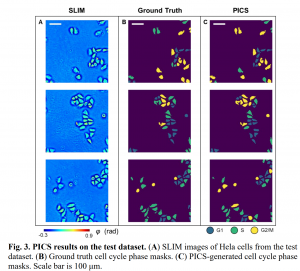
Conventional methods for cell analysis, such as cell viability assay and cell cycle prediction, rely heavily on fluorescence microscopy to monitor nuclei dynamics. These methods inevitably face the typical phototoxicity and photobleaching limitations of fluorescence imaging, which makes them undesirable for rapid, nondestructive and long-term investigation of live cells.Label-free imaging techniques, such as phase imaging, hold a great promise to mitigate these issues. However, label-free images lack specificity of cell nuclei. In this study, by use of deep learning techniques, we investigated effective solutions to extract nuclei dynamics information from phase images for cell analysis. Specifically, three cell analysis tasks were investigated, including cell viability assay, cell cycle detection, and label-free 3D volume and dry mass density rendering. We reformulated these tasks as image-to-image translation problems, and employed EfficientNet-base U-nets to learn end-to-end mappings from phase images to the target images. The target images in the three mentioned tasks are semantic segmentation maps that indicate cell viability at each pixel, semantic segmentation maps that indicate cell cycle at each pixel, and fluorescent images, respectively. The experiment results related to these tasks demonstrate that the proposed solutions hold great promise to perform nondestructive cell analysis in variety of biomedical applications.
Publications
1. Hu, C., He, S., Lee, Y.J., He, Y., Anastasio, M. and Popescu, G., 2021, March. Label-free cell viability assay using phase imagingwith computational specificity (PICS). In Quantitative Phase Imaging VII(Vol. 11653, p. 116531D). International Society for Optics and Photonics.
2. He, Y.R.*, He, S.*, Kandel, M.*, Lee, Y.J., Sobh, N., Anastasio, M. and Popescu, G., 2021, March. Cell cycle detection using phase imaging with computational specificity (PICS). In Quantitative Phase Imaging VII(Vol. 11653, p. 116531R). International Society for Optics and Photonics.
3. Chen, X., Kandel, M., He, S., Lee, Y.J., Sullivan, K., Kong, H.J., Anastasio, M. and Popescu, G., 2021, March. Laser scanning GLIM (LS-GLIM) for label-free imaging of turbid samples. InQuantitative Phase Imaging VII(Vol. 11653, p. 1165304). International Society for Optics and Photonics.
4. Chen, X., Kandel, M.E., He, S., Hu, C., Lee, Y.J., Sullivan, K., Tracy, G., Chung, H.J., Kong, H.J., Anastasio, M. and Popescu, G., 2023. Artificial confocal microscopy for deep label-free imaging. Nature Photonics, pp.1-9.