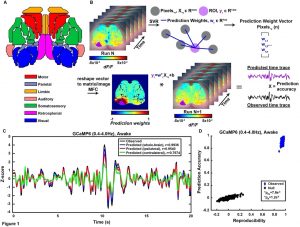
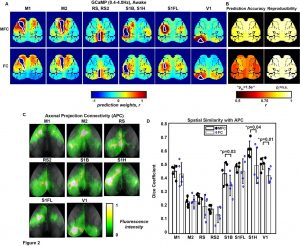
A multivariate functional connectivity approach to mapping brain networks and imputing neural activity in mice
[Paper][Code]
|
|
|
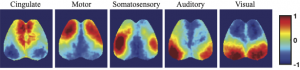
Temporal correlation analysis of spontaneous brain activity (e.g., Pearson “functional connectivity”, FC) has provided insights into the functional organization of the human brain. However, bivariate analysis techniques such as this are often susceptible to confounding physiological processes (e.g., sleep, Mayer-waves, breathing, motion) thus making it difficult to accurately map connectivity in health and disease. In contrast, a multivariate approach to imputing individual neural networks from spontaneous neuroimaging data could be influential to our conceptual understanding of FC and provide performance advantages. Therefore, we analyzed neural calcium imaging data from Thy1-GCaMP6f mice while either awake, asleep, anesthetized, during low and high bouts of motion, or before and after photothrombotic stroke. A linear support vector regression approach was used to determine the optimal weights for integrating the signals from the remaining pixels to accurately predict neural activity in a region of interest (ROI). The resultant weight maps for each ROI were interpreted as multivariate functional connectivity (MFC), resembled anatomical connectivity, and demonstrated a sparser set of strong focused positive connections than traditional FC. While global variations in data have large effects on standard correlation FC analysis, the MFC mapping methods were mostly impervious. Lastly, MFC analysis provided a more powerful connectivity deficit detection following stroke compared to traditional FC.
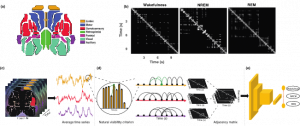
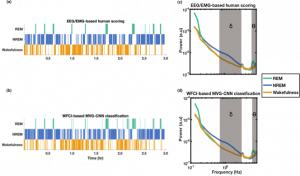
Automated sleep scoring of wide-field optical imaging via multiplex visibility graph and deep learning
[Paper][Code][Dataset]
|
|
|
Wide-field calcium imaging (WFCI) allows for monitoring of cortex-wide neural dynamics in mice. When applied to the study of sleep, WFCI data are manually scored into the sleep states of wakefulness, non-REM (NREM) and REM by use of adjunct EEG and EMG recordings. However, this process is time-consuming and often suffers from low inter- and intra-rater reliability and invasiveness. Therefore, an automated sleep state classification method that operates on WFCI data alone is needed. A hybrid, two-step method is proposed. In the first step, spatial-temporal WFCI data is mapped to multiplex visibility graphs (MVGs). Subsequently, a two-dimensional convolutional neural network (2D CNN) is employed on the MVGs to be classified as wakefulness, NREM and REM. Sleep states were classified with an accuracy of 84% and Cohen’s κ of 0.67. The method was also effectively applied on a binary classification of wakefulness/sleep (accuracy=0.82, κ=0.62) and a four-class wakefulness/sleep/anesthesia/movement classification (accuracy=0.74, κ=0.66). Gradient-weighted class activation maps revealed that the CNN focused on short- and long-term temporal connections of MVGs in a sleep state-specific manner. Sleep state classification performance when using individual brain regions was highest for the posterior area of the cortex and when cortex-wide activity was considered. On a 3-hour WFCI recording, the MVG-CNN achieved a κ of 0.65, comparable to a κ of 0.60 corresponding to the human EEG/EMG-based scoring. The hybrid MVG-CNN method accurately classifies sleep states from WFCI data and will enable future sleep-focused studies with WFCI.
Recurrent autoencoder to identify functional brain networks of spatiotemporal wide-field calcium imaging in mice
[Paper]
|
|
Exploring functional brain networks (FBNs) from wide-field calcium imaging (WFCI) data is important to understand the functional architecture and organization of the brain. In the study, an unsupervised deep learning method is implemented for identifying FBNs from WFCI data. Specifically, a recurrent autoencoder is adapted to extract spatial-temporal latent embeddings of brain activity followed by use of ordinary least square regression to establish the corresponding function brain networks. Spatial similarities are shared between FBNs estimated from learned embeddings and those derived by seed-based correlation method. The proposed method allows investigations about the effect of spatial-temporal calcium dynamics on FBNs.
Publications
-
- Brier, L. M., Zhang, X., Bice, A. R., Gaines, S. H., Landsness, E. C., Lee, J. M., Anastasio, M. A. & Culver, J. P. (2021). A Multivariate Functional Connectivity Approach to Mapping Brain Networks and Imputing Neural Activity in Mice. Cerebral Cortex.
- Landsness, E., & Zhang, X. (2021). Wide-field calcium imaging sleep state database (version 1.0.0). PhysioNet.
- Zhang, X., Landsness, E. C., Chen, W., Miao, H., Tang, M., Brier, L. M., Culver, J. P & Anastasio, M. A. (2021). Automated sleep state classification of wide-field calcium imaging data via multiplex visibility graphs and deep learning. Journal of Neuroscience Methods, 109421.
- Zhang, X., Landsness, E. C., Culver, J. P., & Anastasio, M. A. (2022, March). Identifying functional brain networks from spatial-temporal wide-field calcium imaging data via a recurrent autoencoder. In Neural Imaging and Sensing 2022 (p. PC1194612). SPIE.
- W. Chen, X. Zhang, H. Miao, MJ. Tang, M. A. Anastasio, J. Culver, JM. Lee, EC. Landsness. “Validation of Deep Learning-based Sleep State Classification“. microPublication Biology (2022).
- Zhang, X, Landsness, E. C., Culver J. P., Lee J. M., and Anastasio M. A. . Attention-based CNN-BiLSTM for sleep state classification of spatiotemporal wide-field calcium imaging data. In Neural Imaging and Sensing 2023, vol. 12365, pp. 39-42. SPIE, 2023.